Microbial Symbioses and Diversity Research Experience for Undergraduates (REU)
Contributing Faculty
Arnold, Elizabeth (Betsy)
Research in the Arnold Lab focuses on the ecology and evolution of microbiomes, with a foucs of fungal symbionts of plants and lichens. They use the tools of molecular biology, genomics, and microbiology in fundamental and applied research that addresses how symbioses evolove, how they function in natural conditions, and how they can be used to improve human sustainability, smart agriculture, and the dicscovery of novel pharmaceutical drugs.
Baltrus, David
Research in the Baltrus Lab is focused on understanding molecular dialogues that occur during interactions between bacteria and other organisms. Projects in this lab range from discovering specific genes and proteins that mediate these interactions, uncovering how interactions change over phylogenetic distance, and revealing how these interactions affect evolutionary dynamics within populations and communities. His research team is specifically interested in the role of horizontal gene transfer in introducing novel proteins to mediate these interactions.

Barberán, Albert
The Barberán Lab's research focuses on exploring the links between microbial community patterns, ecosystem functions, global change and landscape management. We use bioinformatics and data analysis tools to understand large-scale dispersal in dust, arid soil ecosystems, invasive plants and soil microbial feedbacks, and the effect of urbanization on microbial communities

Blankinship, Joseph
Research in Dr. Blakinship’s lab focuses on roles of soil microorganisms in controlling ecosystem services, including their glues that help prevent wind and water erosion, their extracellular enzymes that unlock plant nutrients, their residues that sequester carbon, and their metabolic activities that produce and consume atmospheric greenhouse gases. We are working on ways to improve soil health, restore ecosystems, and sustain desert agriculture.

Carini, Paul
Dr. Carini’s team explores the microbial diversity of soil by using cultivation-centric approaches alongside genome analysis. REU students will learn how to culture bacteria from soil using advanced microbiological techniques and identify them using nucleic acid-based molecular tools (PCR). These isolates will become part of a microbial culture collection. A successful 10-week project would include sample collection, microbial isolation and preliminary microbial identification of about 200 soil bacteria.

Cooper, Kerry
The Cooper laboratory research focuses on utilizing different -omic tools, phenotype assays, and animal models to address the genomics, pathogenesis and epidemiology of various bacterial foodborne pathogens including Campylobacter jejuni, Salmonella, Listeria, and Shiga toxin-producing Escherichia coli. Additionally, we are investigating the microbiomes and metagenomics of different agricultural environments, and the impact foodborne pathogens have on the communities, which includes exploring the microbiomes of various fresh produce and how they can be used to prevent colonize by pathogens.

Dhar, Arun
Research in Aquaculture Pathology Laboratory (APL) focuses on discovering pathogens and developing diagnostics for infectious diseases in crustaceans with an emphasis on shrimp. Another area of translational research focus involves understanding the molecular basis of pathogenesis and developing therapies against viral and bacterial diseases in shrimp. Specific REU projects available include:
- Developing molecular tools for shrimp virus detection and evolutionary studies from archived histology samples
- Molecular characterization of a receptor involved in binding binary toxin released by Vibrio parahaemolyticus causing acute hepatopancreatic necrosis disease in shrimp
- Mining pathogen sequence from a host-pathogen sequence pool- an in-silico approach of pathogen discovery.
Duca, Frank
Our lab is interested in the role of the gut-brain axis in the development of obesity and diabetes. We are interested in how dietary and environmental changes, along with shifts in the gut microbiome, alter the ability of the gastrointestinal tract and the brain to regulate glucose and energy homeostasis.

Hunter, Martha (Molly)
Dr. Hunter studies the ecology and evolution of intracellular, maternally inherited bacterial symbionts of insects, and in particular, the effects of symbiont on herbivore and natural enemy biology and reproduction. Current projects focus on studying the fitness effects of the bacterium Rickettsia on whiteflies, genome-enabled research to explore how Cardinium causes cytoplasmic incompatibility in Encarsia wasps, the influence of an environmentally acquired bacterium on the fitness of a stilt bug, and the role of symbionts in interactions between the cochineal scale and its predators.

Maier, Raina
Dr. Maier’s research program focuses on diversity of microorganisms in extreme oligotrophic environments. These living laboratories include the Atacama Desert in Chile, various desert mine tailings sites in the US Southwest, and a carbonate cave (the NSF Kartchner Caverns Microbial Observatory). Energy generation in each of these sites is based to some extent on bacterial/archaeal autotrophy and as such the associated microbial communities and their interactions are of great interest to understanding the ecology of these unusual environments.
Malak, Tfaily
Research in the Tfaily lab revolves around carbon cycling in terrestrial and aquatic ecosystems, the interactions between microbial communities and organic matter, their geochemical environment and the resulting impact on the whole ecosystem. We use a combination of modern analytical molecular techniques (high resolution mass spectrometry, etc.), geochemical (wet chemistry and gas flux) and isotopic techniques (natural abundance, and isotope enrichment) to answer the who, where and how organic matter degradation and formation takes place in different ecosystems.
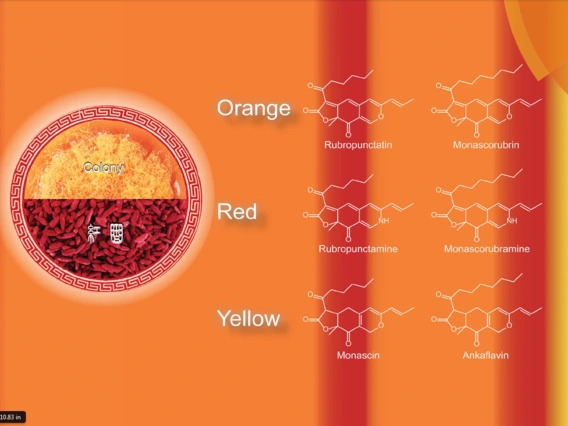
Molnar, Istvan
Dr. Molnar’s research is centered on the study of secondary metabolites that microorganisms (bacteria, fungi, and algae) produce, primarily as effectors of symbiotic interactions. His studies are designed to unravel the biosynthesis of these compounds by isolating the gene clusters that encode the necessary biosynthetic machinery. His lab is also working on using such knowledge to re-engineer the biosynthetic genes and gene clusters, and thereby, produce altered molecules with improved biological activities for human and agricultural use.

Pryor, Barry
Dr. Pryor’s program is focused on understanding the functional and evolutionary interactions of fungi with their environment. Areas of interest include mechanisms of pathogenesis and management of fungal diseases of field and vegetable crops, and evolution and speciation of plant pathogenic fungi. Additional interests include fungal community composition in native and agricultural ecosystems, particularly those in semi-arid and arid zones.
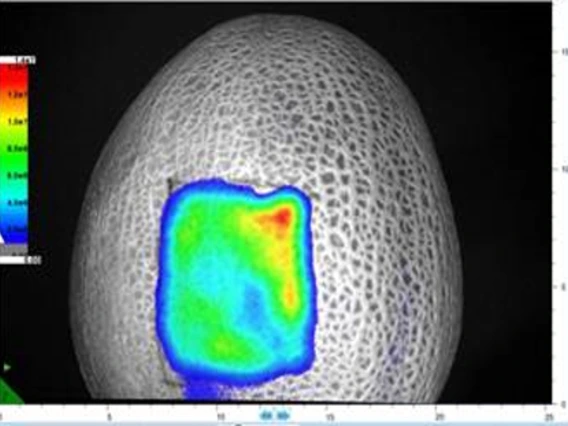
Ravishankar, Sadhana
The Ravishankar’s lab studies foodborne pathogenic bacteria including antibiotic resistant strains using various technologies and multiple hurdle approach. Another focus is on natural antimicrobials and their applications in organic foods, plant antimicrobial washes for fresh produce, antimicrobial and anti-oxidative activities of plant compounds; Bacterial attachment and biofilm formation in food production environments; Survival and prevalence of foodborne pathogens in fresh produce growing environments/organic composts; Stress tolerance responses of foodborne pathogenic bacteria.
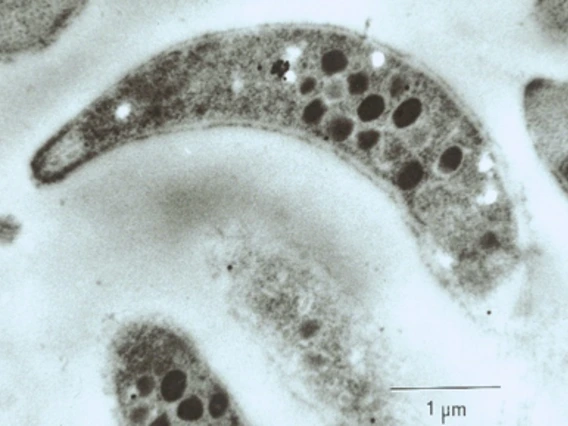
Riggs, Michael
The Riggs lab research center on the immunobiology and molecular pathogenesis of parasitic protozoal diseases of zoonotic importance. Current research is focused on development of recombinant and synthetic vaccines for cryptosporidiosis; immunotherapy and new drug discovery for cryptosporidiosis; definition of the molecular pathogenesis of host cell recognition, attachment, and invasion by Cryptosporidium; structural characterization of Cryptosporidium glycoprotein ligands; animal model development; and improved methods for diagnosis and detection.
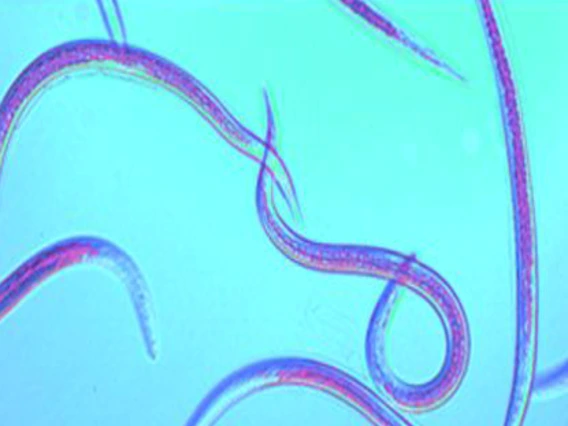
Stock, S. Patricia
Dr. Stock’s research program focuses on the study of nematode-bacteria symbiosis as model systems. Specifically, her research centers on insect pathogenic nematodes (Steinernema and Heterorhabditis spp.) and their bacterial symbiont bacteria (Xenorhabdus and Photorhabdus spp.) which represent an emerging model of terrestrial animal-microbe symbiotic relationships. Current projects available in her lab include:
- Mechanisms involved in the maintenance and transmission of nematode bacterial symbionts;
- How partner specificity and fidelity is maintained;
- Assessment of novel bacterial symbiont bioactive molecules with insecticidal, nematicidal, antibiotic, and antimycotic properties.
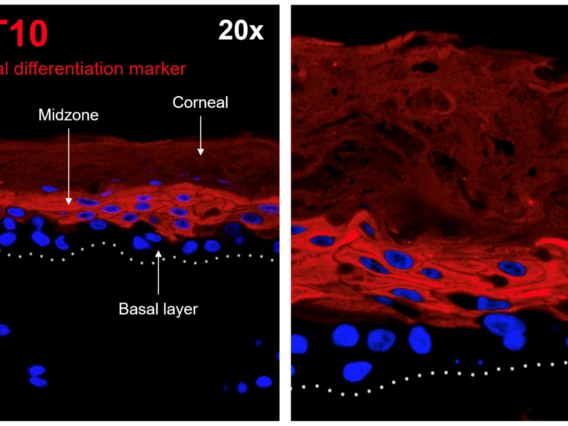
Van Doorslaer, Koenraad
The Van Doorslaer lab is interested in host-pathogen interactions. Specifically, we study how viruses infect and establish longterm infections in the host. We study how these interactions can lead to cancer progression. We leverage state of the art tissue culture methods with genomics and bioinformatics to understand the evolutionary interactions of viruses with their hosts. While we primarily focus on papillomaviruses, we recently started studying the SARS-2-CoV virus. Potential projects in the lab include
1) Discovery, description, and evolutionary analyses of novel animal viruses
2) Generation of recombinant HPV16 genomes
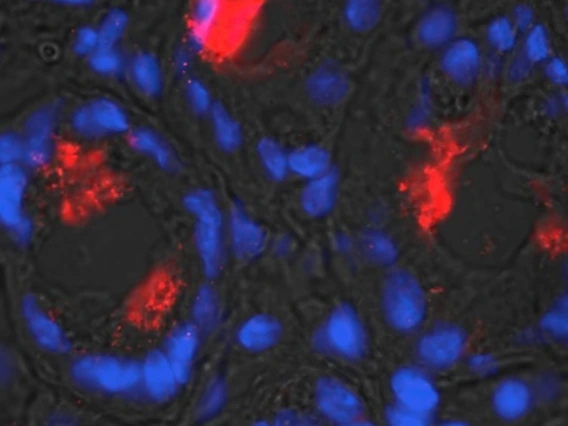
Vedantam, Gayatri
Dr. Vedantam’s research is dedicated to the concept of One-Health – a unified approach to addressing health issues common to humans, animals and agriculture. Projects currently underway include those focused on the bacterial pathogen Clostridium difficile. Specific REU projects available in his lab include:
- Molecular typing of new Clostridium difficile strains;
- Generation of C. difficile strain gene libraries for phenotype screening;
- Interrogation of host-cell signaling networks during C. difficile attachment.